Detecting Bad Data and Anomalies with the TDDA Library (Part I)
Posted on Fri 04 May 2018 in TDDA
The test-driven data analysis library, tdda, has two main kinds of functionality
- support for testing complex analytical processes
with
unittestorpytest - support for verifying data against constraints, and optionally for discovering such constraints from example data.
Until now, however, the verification process has only reported which constraints failed to be satisfied by a dataset.
We have now extended the tdda library to allow identification of
individual failing records, allowing it to act as a general purpose
anomaly detection framework.
The new functionality is available through a new detect_df API call,
and from the command line with the new tdda detect command.
The diagram shows conceptually how detection works, separating out anomalous records from the rest.
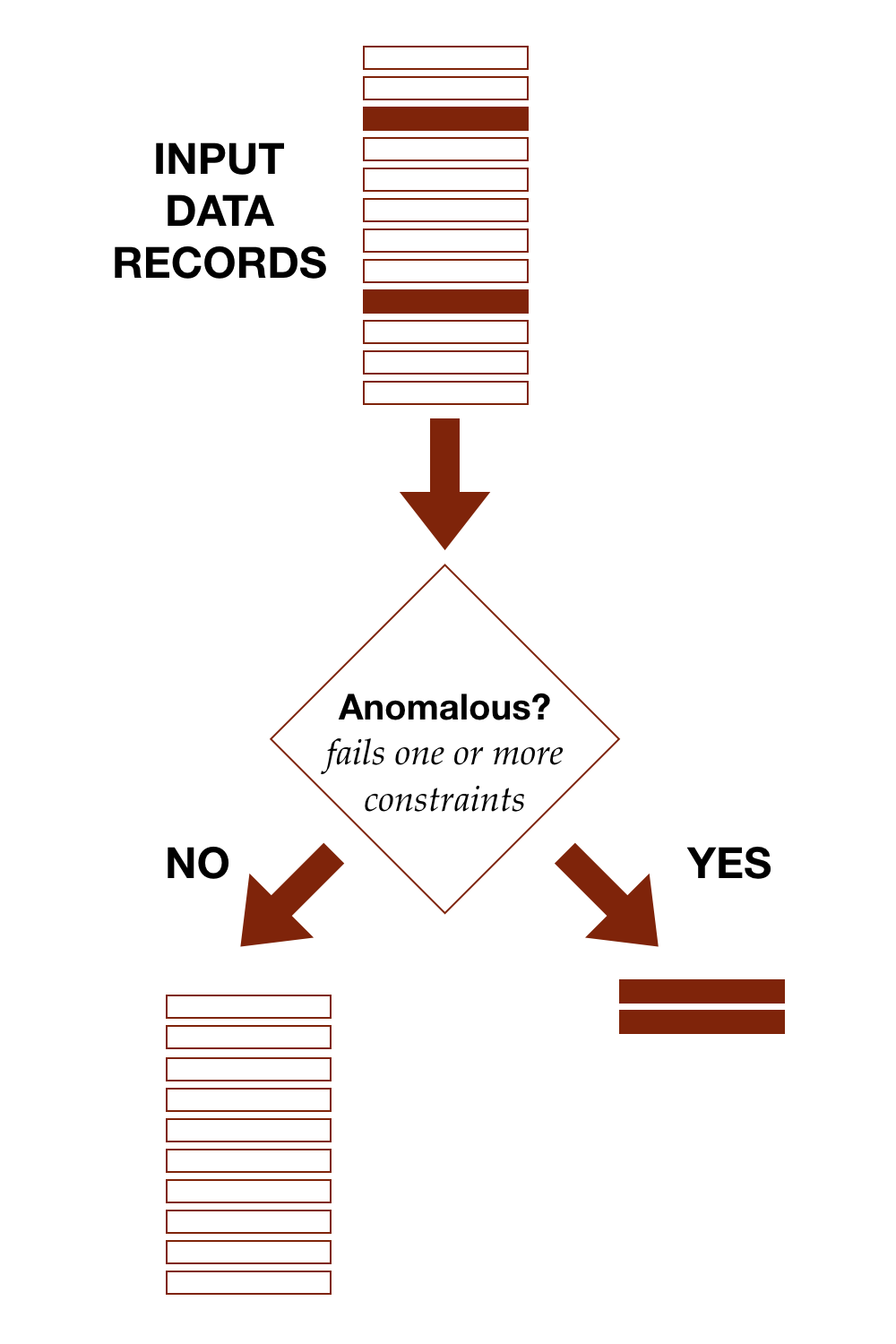
With the TDDA framework, anomalous simply means fails at least one constraint. We'll discuss cases in which the constraints have been developed to try to model some subset of data of interest (defects, high-risk applicants, heart arrythmias, flawless diamonds, model patients etc.) in part II of this post. In those cases, we start to be able to discuss classifications such as true and false positives, and true and false negatives.
Example Usage from the Command Line
Suppose we have a simple transaction stream with just three fields,
id, category and price, like this:
id category price
710316821 QT 150.39
516025643 AA 346.69
414345845 QT 205.83
590179892 CB 55.61
117687080 QT 142.03
684803436 AA 152.10
611205703 QT 39.65
399848408 AA 455.67
289394404 AA 102.61
863476710 AA 297.82
534170200 KA 80.96
898969231 QT 81.39
255672456 QT 71.67
133111344 TB 229.19
763476994 CB 338.40
769595502 QT 310.19
464477044 QT 54.41
675155634 QT 199.07
483511995 QT 209.53
416094320 QT 83.31
and the following constraints (which might have been created by hand,
or generated using the tdda discover command).
-
id(integer): Identifier for item. Should not be null, and should be unique in the table -
category(string): Should be one of “AA”, “CB”, “QT”, “KA” or “TB” -
price(floating point value): unit price in pounds sterling. Should be non-negative and no more than 1,000.00.
This would be represented in a TDDA file with the following constraints.
{
"fields": {
"id": {
"type": "int",
"max_nulls": 0,
"no_duplicates": true
},
"category": {
"type": "string",
"max_nulls": 0,
"allowed_values":
["AA", "CB", "QT", "KA", "TB"]
},
"price": {
"type": "real",
"min": 0.0,
"max": 1000.0,
"max_nulls": 0
}
}
}
We can use the tdda verify command to verify a CSV file or a feather
file1 against these files, and get a summary of which constraints pass
and fail. If our data is in the file items.feather and the JSON
constraints are in constraints.tdda, and there are some violations
we will get output exemplified by the following:
$ tdda verify items.feather constraints.tdda
FIELDS:
id: 1 failure 2 passes type ✓ max_nulls ✓ no_duplicates ✗
category: 1 failure 2 passes type ✓ max_nulls ✓ allowed_values ✗
price: 2 failures 2 passes type ✓ min ✓ max ✗ max_nulls ✗
SUMMARY:
Constraints passing: 6
Constraints failing: 4
The new tdda detect command allows us to go further and find which
individual records fail.
We can use the following command to write out a CSV file, bads.csv,
containing the records that fail constraints:
$ tdda detect items.feather constraints.tdda bads.csv --per-constraint --output-fields
The flag --per-constraint tells the software to write out a boolean
column for each constraint, indicating whether the record
passed, and the --output-fields tells the software to include all
the input fields in the output.
The result is the following CSV file:
id,category,price,id_nodups_ok,category_values_ok,price_max_ok,price_nonnull_ok,n_failures
113791348,TQ,318.63,true,false,true,true,1
102829374,AA,65.24,false,true,true,true,1
720313295,TB,1004.72,true,true,false,true,1
384044032,QT,478.65,false,true,true,true,1
602948968,TB,209.31,false,true,true,true,1
105983384,AA,8.95,false,true,true,true,1
444140832,QT,1132.87,true,true,false,true,1
593548725,AA,282.58,false,true,true,true,1
545398672,QT,1026.4,true,true,false,true,1
759425162,CB,1052.72,true,true,false,true,1
452691252,AA,1028.19,true,true,false,true,1
105983384,QT,242.64,false,true,true,true,1
102829374,KA,71.64,false,true,true,true,1
105983384,AA,10.24,false,true,true,true,1
405321922,QT,85.23,false,true,true,true,1
102829374,,100000.0,false,false,false,true,3
872018391,QT,51.69,false,true,true,true,1
862101984,QT,158.53,false,true,true,true,1
274332319,AA,1069.25,true,true,false,true,1
827919239,QT,1013.0,true,true,false,true,1
105983384,QT,450.68,false,true,true,true,1
102829374,,100000.0,false,false,false,true,3
872018391,QT,199.37,false,true,true,true,1
602948968,KA,558.73,false,true,true,true,1
328073211,CB,1031.67,true,true,false,true,1
405321922,TB,330.97,false,true,true,true,1
334193154,QT,1032.31,true,true,false,true,1
194125540,TB,,true,true,,false,1
724692620,TB,1025.81,true,true,false,true,1
862101984,QT,186.76,false,true,true,true,1
593548725,QT,196.56,false,true,true,true,1
384044032,AA,157.25,false,true,true,true,1
which, we can read a bit more easily if we reformat this (using · to denote nulls) as:
id category price id category price price n_failures
_nodups _values _max _nonnull
_ok _ok _ok _ok
113791348 TQ 318.63 true false true true 1
102829374 AA 65.24 false true true true 1
720313295 TB 1,004.72 true true false true 1
384044032 QT 478.65 false true true true 1
602948968 TB 209.31 false true true true 1
105983384 AA 8.95 false true true true 1
444140832 QT 1132.87 true true false true 1
593548725 AA 282.58 false true true true 1
545398672 QT 1,026.40 true true false true 1
759425162 CB 1,052.72 true true false true 1
452691252 AA 1,028.19 true true false true 1
105983384 QT 242.64 false true true true 1
102829374 KA 71.64 false true true true 1
105983384 AA 10.24 false true true true 1
405321922 QT 85.23 false true true true 1
102829374 · 100,000.00 false false false true 3
872018391 QT 51.69 false true true true 1
862101984 QT 158.53 false true true true 1
274332319 AA 1,069.25 true true false true 1
827919239 QT 1,013.00 true true false true 1
105983384 QT 450.68 false true true true 1
102829374 · 100,000.00 false false false true 3
872018391 QT 199.37 false true true true 1
602948968 KA 558.73 false true true true 1
328073211 CB 1,031.67 true true false true 1
405321922 TB 330.97 false true true true 1
334193154 QT 1,032.31 true true false true 1
194125540 TB · true true · false 1
724692620 TB 1,025.81 true true false true 1
862101984 QT 186.76 false true true true 1
593548725 QT 196.56 false true true true 1
384044032 AA 157.25 false true true true 1
Command Line Syntax
The basic form of the command-line command is:
$ tdda detect INPUT CONSTRAINTS OUTPUT
where
INPUTis normally either a.csvfile, in a suitable format, or a.featherfile1 containing a DataFrame, preferably with an accompanying.pmmfile1CONSTRAINTSis a JSON file containing constraints, usually with a.tddasuffix. This can be created by thetdda discovercommand, or edited by hand.OUTPUTis again either a.csvor.featherfile to be created with the output rows. If thepmmiflibrary is installed, a.pmmmetadata file will be generated alongside the.featherfile, when.featheroutput is requested.
Options
Several command line options are available to control the detailed behaviour:
-
defaults: If no command-line options are supplied:
- only failing records will be written
- only a record identifier and the number of failing constraints
will be written.
- When the input is Pandas, the record identifier will be the index for the failing records;
- when the input is a CSV file, the record identifier will be the row number, with the first row after the header being numbered 1.
-
--per-constraintwhen this is added, an_okcolumn will also be written for every constraint that has any failures, withtruefor rows that satisfy the contraint,falsefor rows that do not satisfy the constraint and a missing value where the constraint is inapplicable (which does not count as a failure). -
--output-fields [FIELD1 FIELD2 ...]If the--output-fieldsflag is used without specifying any fields, all fields from the input will be included in the output. Alternatively, a space-separated list of fields may be provided, in which case only those will be included. Whenever this option is used, no index or row-number is written unless specifically requested -
--write-allIf this flag is used, all records from the input will be included in the output, including those that have no constraint failures. -
--indexThis flag forces the writing of the index (for DataFrame inputs) or row number (for CSV inputs). -
--intWhen writing boolean values to CSV files (either from input data or as per-constraint output fields), use1fortrueand0forfalse.
API Access
The detection functionality is also available through the TDDA library's
API with a new detect_df function, which takes similar parameters
to the command line. The corresponding call, with a DataFrame df in
memory, would be:
from tdda.constraints import detect_df
verification = detect_df(df, 'constraints.tdda', per_constraint=True,
output_fields=[])
bads_df = verification.detected()
-
The feather file format is an interoperable way to save DataFrames from Pandas or R. Its aim is to preserve metadata better and be faster than CSV files. It has a few issues, particularly around types and nulls, and when available, we save a secondary
.pmmfile alongside.featherfiles which makes reading and writing them more robust when our extensions in thepmmiflibrary are used. We'll do a future blog post about this, but if you install bothfeatherandpmmifwithpip install featherandpip install pmmif, and usefeatherpmm.write_dataframe, imported frompmmif, rather thanfeather.write_dataframe, you should get more robust behaviour. ↩↩↩