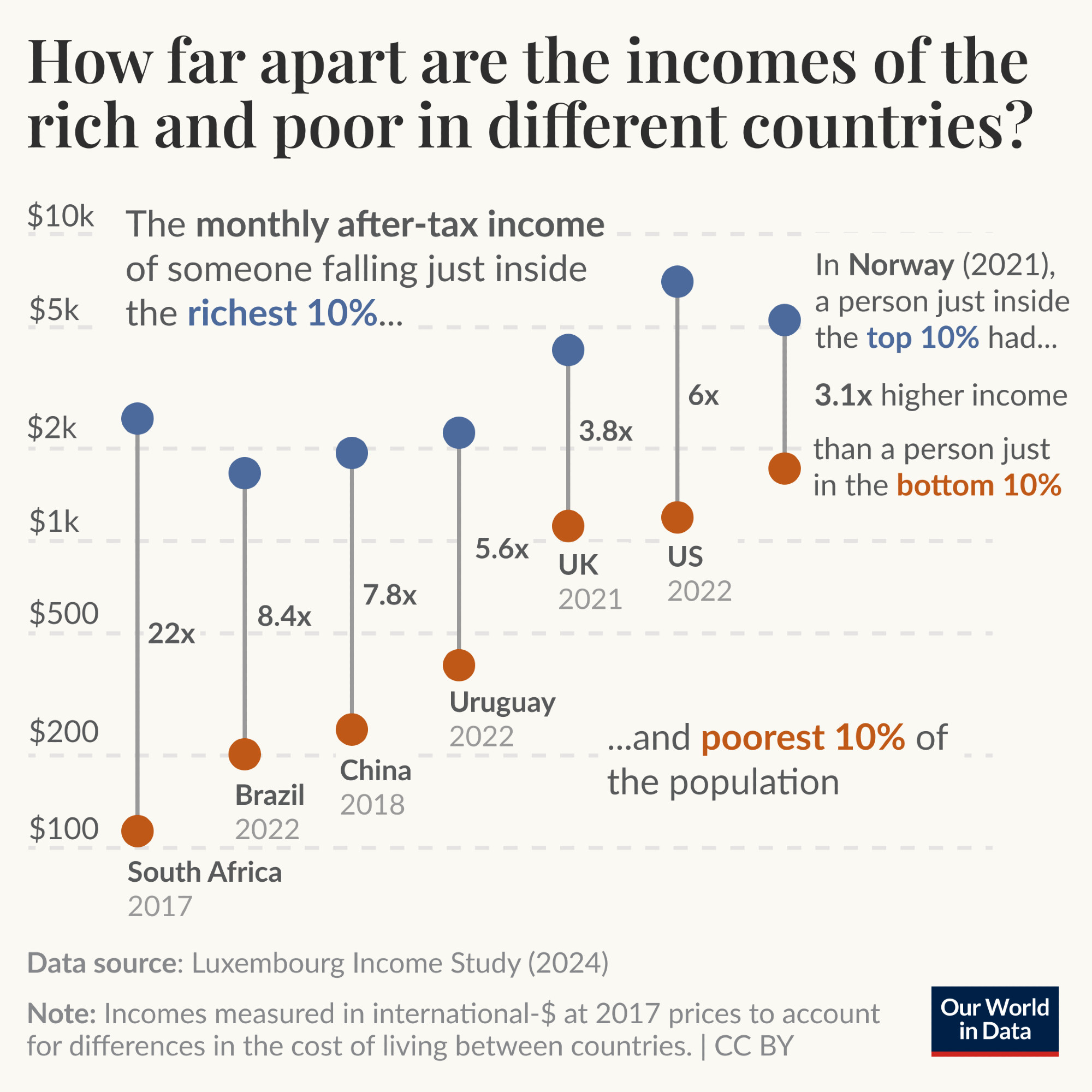
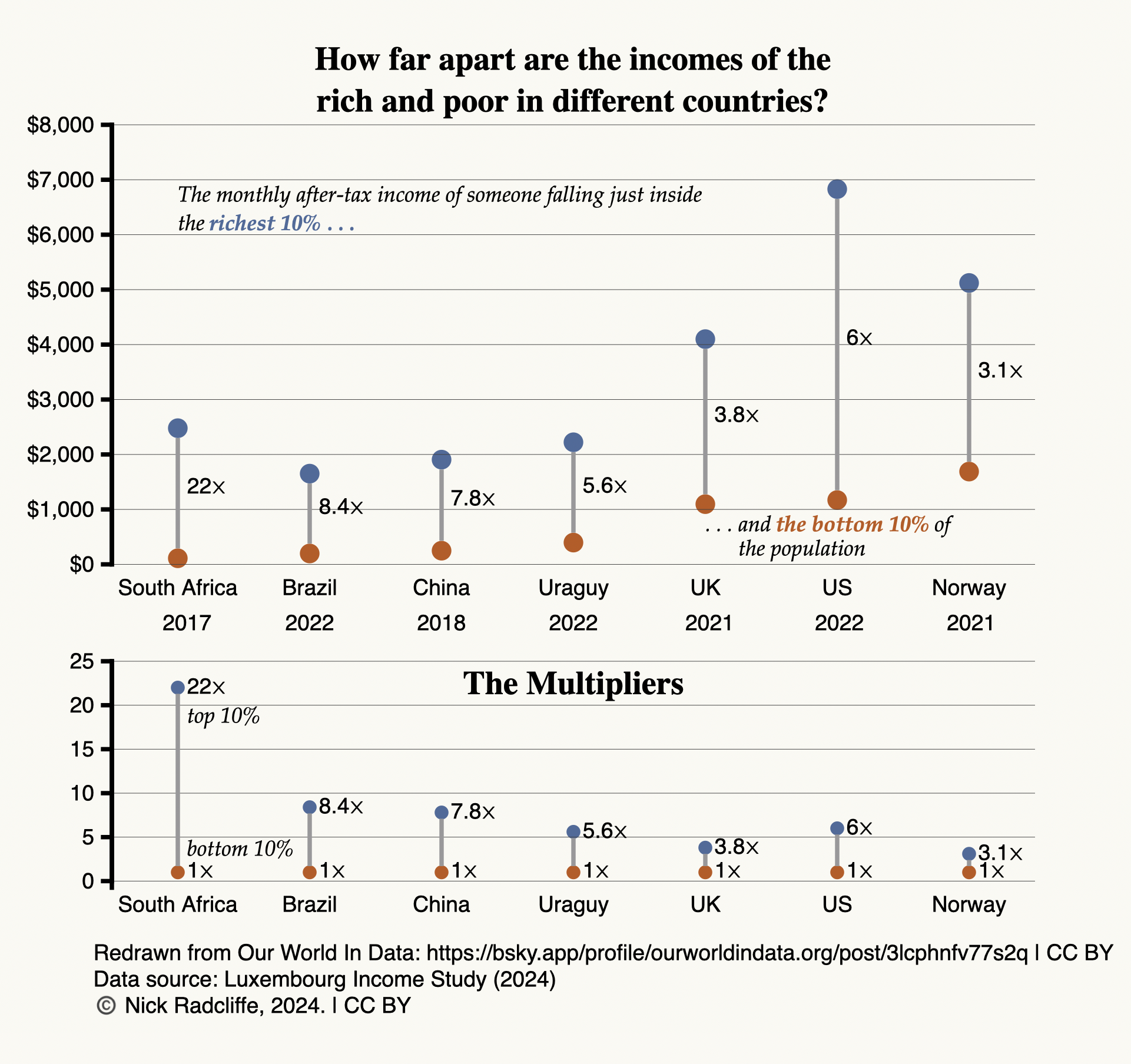
tdda.serial: Metadata for Flat Files (CSV Files)
Posted on Mon 23 June 2025 in misc
Almost all data scientists and data engineers have to work with flat files (CSV files) from time to time. Despite their many problems, CSVs are too ubiquitous, too universal, and (whisper it) have too many strengths for them to be likely to disappear. Even if they did, they would quickly be reinvented. The problems with them are widely known and discussed, and will be familar to almost everyone who works with them. They include issues with encodings, types, quoting, nulls, headers, and with dates and times. My favourite summary of them remains Jesse Donat's Falsehoods Programmers Believe about CSVs. I wrote about them on this blog nearly four years ago (Flat Files).
Over the last year or so I've been writing a book on test-driven data analysis. The only remaining chapter without a full draft discusses the same topics as this post—metadata for CSV files and new parts of the TDDA software that assist with its creation and use. This post documents my current thinking, plans and ambitions in this area, and shows some of what is already implemented.1
A Metadata Format for Flat Files: tdda.serial
The core of the new work is a new format, tdda.serial, for describing
data in CSV files.
The previous post showed an example (“XMD”) metadata file used by the Miró software from my company Stochastic Solutions, which was as follows:2
<?xml version="1.0" encoding="UTF-8"?>
<dataformat>
<sep>,</sep> <!-- field separator -->
<null></null> <!-- NULL marker -->
<quoteChar>"</quoteChar> <!-- Quotation mark -->
<encoding>UTF-8</encoding> <!-- any python coding name -->
<allowApos>True</allowApos> <!-- allow apostophes in strings -->
<skipHeader>False</skipHeader> <!-- ignore the first line of file -->
<pc>False</pc> <!-- Convert 1.2% to 0.012 etc. -->
<excel>False</excel> <!-- pad short lines with NULLs -->
<dateFormat>eurodt</dateFormat> <!-- Miró date format name -->
<fields>
<field extname="mc id" name="ID" type="string"/>
<field extname="mc nm" name="MachineName" type="int"/>
<field extname="secs" name="TimeToManufacture" type="real"/>
<field extname="commission date" name="DateOfCommission"
type="date"/>
<field extname="mc cp" name="Completion Time" type="date"
format="rdt"/>
<field extname="sh dt" name="ShipDate" type="date" format="rd"/>
<field extname="qa passed?" name="Passed QA" type="bool"/>
</fields>
<requireAllFields>False</requireAllFields>
<banExtraFields>False</banExtraFields>
</dataformat>
Here is one equivalent way of expressing essentially the same information
in the (evolving) tdda.serial format:
{
"format": "http://tdda.info/ns/tdda.serial",
"writer": "tdda.serial-2.2.15",
"tdda.serial": {
"encoding": "UTF-8",
"delimiter": "|",
"quote_char": "\"",
"escape_char": "\\",
"stutter_quotes": false,
"null_indicators": "",
"accept_percentages_as_floats": false,
"header_row_count": 1,
"map_missing_trailing_cols_to_null": false,
"fields": {
"mc id": {
"name": "ID",
"fieldtype": "int"
},
"mc nm": {
"name": "Name",
"fieldtype": "string"
},
"secs": {
"name": "TimeToManufacture",
"fieldtype": "int"
},
"commission date": {
"name": "DateOfCommission",
"fieldtype": "date",
"format": "iso8601date"
},
"mc cp": {
"name": "CompletionTime",
"fieldtype": "datetime",
"format": "iso8601datetime"
},
"sh dt": {
"name": "ShipDate",
"fieldtype": "date",
"format": "iso8601date"
},
"qa passed?": {
"name": "PassedQA",
"fieldtype": "bool",
"true_values": "yes",
"false_values": "no"
}
}
}
}
The details don't matter too much at this stage, and may yet change,
but briefly here we see the file
(typically with a .serial extension), describing:
- the text encoding used for the data (
UTF-8); - the field separator (pipe,
|); - the quote character (double quote,
"); - the escape character (
\), which is used to escape double quotes in double-quoted strings, among other things; - whether quotes are stuttered or escaped within quoted strings;
- the string used to denote null values (this can be a single string or a list);
- the number of header rows;
- an explicit note not to accept percentages in the file as floating-point values;
- whether or not lines with too few fields should be regarded as having nulls for the apparently missing fields. (Excel usually does not write values after the last non-empty cell in each row on a worksheet.)
- information about individual fields.
In this case, a dictionary is used to map names in the flat file
to names to be used in the dataset. Numbers can also be used
to indicate column position, particularly if there is no header,
though they have to be quoted because this is JSON.
Field types are also specified, together with any extra information
required, e.g. the non-standard true and false values for the
boolean field
collected?(in the file), which becomesHasBeenCollectedonce read. Formats for the date and time fields are also specified here.
When the fields are presented as a dictionary, as here, this allows
for the possibility that there are other fields in the file, for
which metadata is not provided. If a list is used instead, the
field list is taken to be complete. In this case, external names
can be provided using an csvname attribute, if they are different.
Pretty much everything is optional, and, where appropriate,
defaults can be put in the main section and over-ridden on
a per-field basis. This is useful if, for example, one or two fields
use different null markers from the default, or if multiple date formats
are used. (The format key will probably change to dateformat and
boolformat to make this overriding work better.)
Here is a simple example of its use with Pandas.
Suppose we have the following pipe-separated flat file,
with the name machines.psv.
mc id|mc nm|secs|commission date|mc cp|sh dt|qa passed?
1111111|"Machine 1"|86400|2025-06-01|2025-06-07T12:34:56|2025-06-21|yes
2222222|"Machine 2"||2025-06-02|2025-06-08T12:34:57|2025-06-22
3333333|"Machine 3"|86399|2025-06-03|2025-06-09T12:34:55|2025-06-22|no
Then we can use the following Python code to load the data,
informed by the metadata in machines.serial (the example
shown above).
from tdda.serial import csv_to_pandas
df = csv_to_pandas('machines.psv', 'machines.serial')
print(df, '\n')
df.info()
This produces the following output:
$ python pd-read-machines.py
ID Name TimeToManufacture DateOfCommission CompletionTime ShipDate PassedQA
0 1111111 Machine 1 86400 2025-06-01 2025-06-07 12:34:56 2025-06-21 True
1 2222222 Machine 2 <NA> 2025-06-02 2025-06-08 12:34:57 2025-06-22 <NA>
2 3333333 Machine 3 86399 2025-06-03 2025-06-09 12:34:55 2025-06-22 False
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 3 entries, 0 to 2
Data columns (total 7 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 ID 3 non-null Int64
1 Name 3 non-null string
2 TimeToManufacture 2 non-null Int64
3 DateOfCommission 3 non-null datetime64[ns]
4 CompletionTime 3 non-null datetime64[ns]
5 ShipDate 3 non-null datetime64[ns]
6 PassedQA 2 non-null boolean
dtypes: Int64(2), boolean(1), datetime64[ns](3), string(1)
memory usage: 288.0 bytes
There's nothing particularly special here, but Pandas has read the file correctly using the metadata to understand
- the pipe separator;
- the date and time formats;
- the
yes/noformat ofPassedQA; - the null indicator;
- the intended, more usable internal field names;
- field types, here defaulting to nullable types.
As with the pandas.read_csv, we can choose whether to prefer
nullable types, but the default using tdda.serial is to do so.
In this case, the date formats and null indicators would be fine anyway,
with Pandas defaults, but here we could instead have specified, say,
European dates and ? for nulls.
This code:
from tdda.serial load_metadata, serial_to_pandas_read_csv_args
from rich import print as rprint
md = load_metadata('machines.serial')
kwargs = serial_to_pandas_read_csv_args(md)
rprint(kwargs)
shows the parameters actually passed to pandas.read_csv:
{
'dtype': {'ID': 'Int64', 'Name': 'string', 'TimeToManufacture': 'Int64', 'PassedQA': 'boolean'},
'date_format': {'DateOfCommission': 'ISO8601', 'CompletionTime': 'ISO8601', 'ShipDate': 'ISO8601'},
'parse_dates': ['DateOfCommission', 'CompletionTime', 'ShipDate'],
'sep': '|',
'encoding': 'UTF-8',
'escapechar': '\\',
'quotechar': '"',
'doublequote': False,
'na_values': [''],
'keep_default_na': False,
'names': ['ID', 'Name', 'TimeToManufacture', 'DateOfCommission', 'CompletionTime', 'ShipDate', 'PassedQA'],
'header': 0,
'true_values': ['yes'],
'false_values': ['no']
}
We can do the very similar things using Polars (and “soon”, other libraries). Here's a way to read the file with Polars:
from tdda.serial import csv_to_polars
df = csv_to_polars('machines.psv', 'machines.serial',
map_other_bools_to_string=True)
print(df)
which produces:
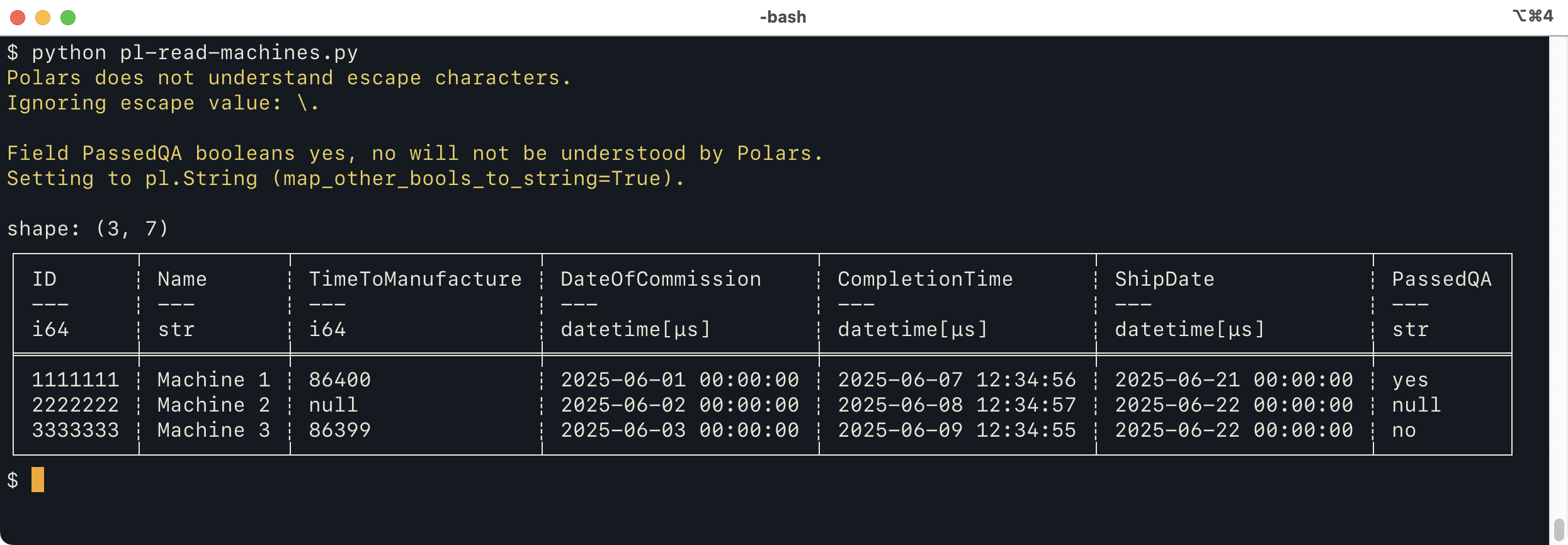
This does mostly the same thing as the Pandas version, but
issues two warnings. The first is because an
escape character is specified, which the Polars CSV reader
doesn't really understand.
The second warning is because the Polars CSV reader can't handle non-standard
booleans. By default, when these are specified for Polars,
tdda.serial will issue a warning but still call polars.read_csv
to read the file, because they might not, in fact, be used.
The parameter passed in the Python code above
(map_other_bools_to_string=True) tells tdda.serial to direct Polars to read
this column as a string instead (as it would if we didn't specify a type).
Of course, it would be possible to have the reader then go through and
turn the strings into booleans after reading, but that feels like
more a metadata library should do.
The warnings helpfully tell you what to look out for as possible issues when the file is read. This as an example of a principle I'm trying to use throughout tdda.serial: when there's something in the serial metadata that a given reader might not be able to handle correctly, issue a warning, and possibly provide an option to control that behaviour.
We can do the same thing as we did for Pandas and look at the arguments generated for Polars, using the following, very similar, Python code:
from tdda.serial import (load_metadata, serial_to_polars_read_csv_args)
from rich import print as rprint
md = load_metadata('machines.serial')
kwargs = serial_to_polars_read_csv_args(md, map_other_bools_to_string=True)
rprint(kwargs)
This produces
{
'separator': '|',
'quote_char': '"',
'null_values': [''],
'encoding': 'UTF-8',
'schema': {
'ID': Int64,
'Name': String,
'TimeToManufacture': Int64,
'DateOfCommission': Datetime,
'CompletionTime': Datetime,
'ShipDate': Datetime,
'PassedQA': String
},
'new_columns': [
'ID',
'Name',
'TimeToManufacture',
'DateOfCommission',
'CompletionTime',
'ShipDate',
'PassedQA'
]
}
The only subtlety here is that the types in Schema are actual polars types
(pl.Int64 etc.) rather than strings, hence their not being quoted.
(They're not prefixed because repr(pl.Int64) is the string "Int64",
which prints as Int64.)
The library can also write a tdda.serial file containing the Polars
arguments explicitly. It looks like this:
{
"format": "http://tdda.info/ns/tdda.serial",
"writer": "tdda.serial-2.2.15",
"polars.read_csv": {
"separator": "|",
"quote_char": "\"",
"null_values": [
""
],
"encoding": "UTF-8",
"schema": {
"ID": "Int64",
"Name": "String",
"TimeToManufacture": "Int64",
"DateOfCommission": "Datetime",
"CompletionTime": "Datetime",
"ShipDate": "Datetime",
"PassedQA": "String"
},
"new_columns": [
"ID",
"Name",
"TimeToManufacture",
"DateOfCommission",
"CompletionTime",
"ShipDate",
"PassedQA"
]
}
}
Here, because we need to serialize the tdda.serial file as JSON, the
polars types are mapped to their string names. The tdda library
takes care of the conversion in both directions.
A single .serial file can contain multiple flavours of
metadata—tdda.serial, polars.read_csv, pandas.read_csv etc.
When it does, a call to load_metadata can specify a preferred flavour,
or let the library choose. My hope, however, is that in most cases
the tdda.serial section will contain enough information to work
as well as a library-specific specification.
Goals for tdda.serial
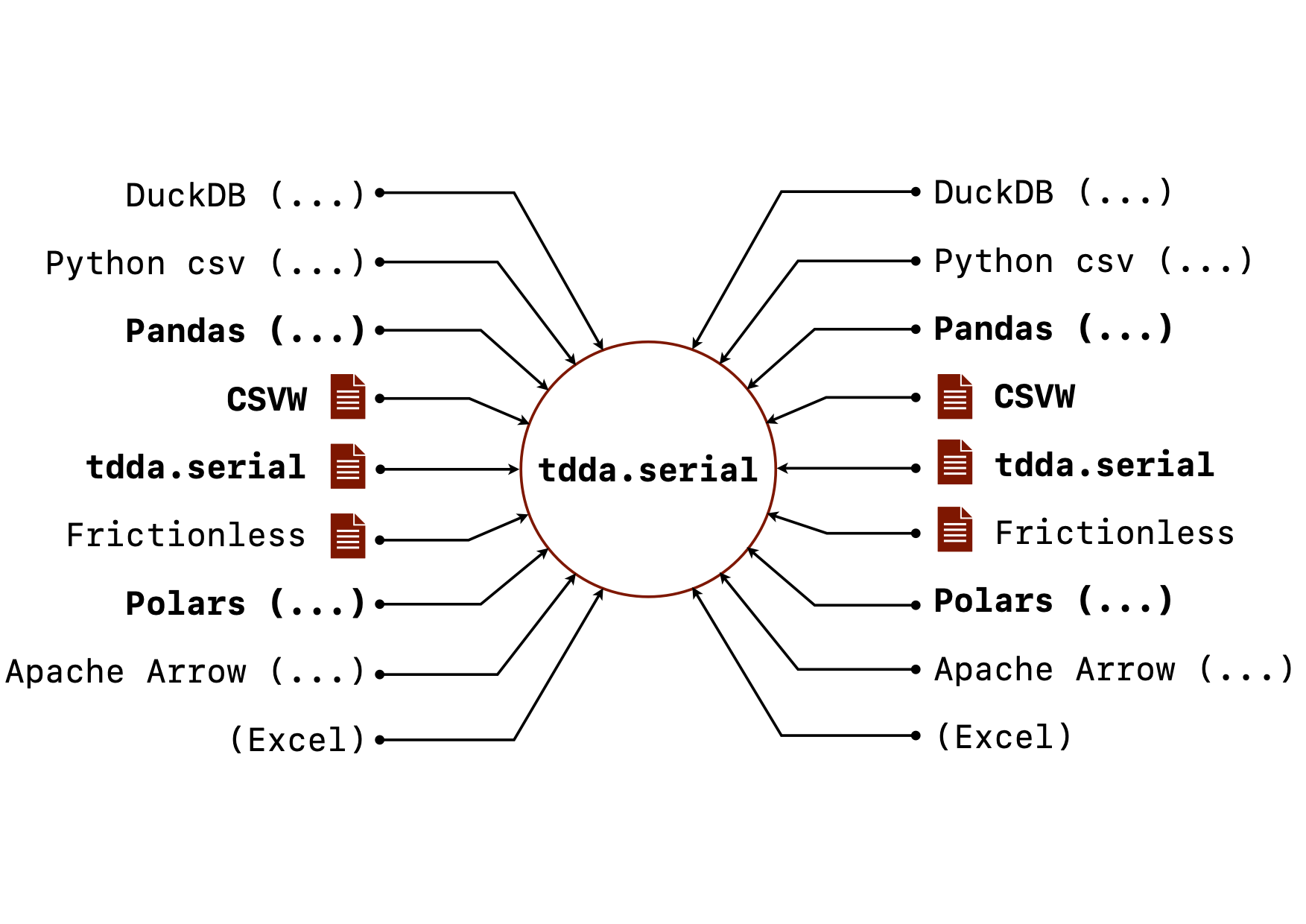
When I went to write down the goals for tdda.serial, I was surprised
at how long the list was. Not all of this is implemented but here is the
current state of the goals for tdda.serial. (The image above shows
the vision for it, with the bold parts mostly implemented, and the
rest currently only planned.)
- Describe Flat File Formats.
Allow accurate representation, full or partial, of flat-file formats
used (or potentially used) by one or more concrete flat files.
or .
- It primarily targets comma-separated values (
.csv) and related formats (tab-separated, pipe-separated etc.), but also potentially other tabular data. It could, for example, be used to describe things like date formats and numeric subtypes for tabular data stored in JSON or JSON Lines. - Full or partial is important. When reading data, it is often convenient only to specify things that are causing problems. On write, fuller specifications are, of course, desirable.
- It primarily targets comma-separated values (
- Read Flat Files.
Assist with reading flat files correctly, based on metadata in
.serialfiles and other formats (like CSVW), primarily using data in the"tdda.serial"format.- Convert metadata currently stored as
tdda.serialto dictionaries of arguments for other libraries that work with CSVs. - Provide an API to get such libraries to read flat-file data correctly, guided by the metadata
- Generate code to get such libraries to read flat-file data correctly, guided by the metadata. Assist with writing flat files in documented formats.
- Interoperate, where possible, with other metadata formats like CSVW and Frictionless.
- Convert metadata currently stored as
- Generate tdda.serial Metadata Files. Assist with generating metadata describing the format of CSV files based on the write arguments provided to the writing software.
- Write Flat Files.
Assist with getting libraries to write CSV files using a format specified
in a
tdda.serialfile.- This provides a second way of increasing interoperability: we can help readers to read from a specific format, and writers to write to that same format.
- Assist/Support other Software Reading, Writing, and otherwise
handling Flat Files.
- DataFrame Libraries
- Pandas
- Polars
- Apache Arrow
- Databases
- DuckDB
- SQLite
- Postgres
- MySQL
- Miscellaneous
- Python
csv - tdda
- Python
- DataFrame Libraries
- Support Library-specific Read/Write Metadata.
Provide a mechanism for documenting library-specific read/write
parameters for CSV files explicitly:
- For storing the library-specific write parameters used with
pandas.to_csv,polars.write_csvin.serialfiles (and the ability to use such parameters) - For storing the library-specific read parameters required to
read a flat file with high fidelity using,
e.g.
pandas.read_csv,polars.read_csvetc.
- For storing the library-specific write parameters used with
- Assist with Format Choice. Provide a mechanism for helping to choose a good CSV format for a concrete dataset to be written, e.g. choosing null indicators that are not likely to be confused with serialized non-null values in the dataset.
- SERDE Verification. Provide mechanisms for checking whether a dataset can be round-tripped successfully to a flat file (i.e. that the same library, at least, can write data to a flat file, read it back, and recover identical, equivalent, or similar data).3
-
CLI Tools. Through the associated command-line tool,
tdda diff, and equivalent API functions, to check whether two datasets are equivalent.- In the case of the command-line tool this is two datasets on
disk (flat files, parquet files etc.). It might also be possible
to compare two database tables, in the same or different RDBMS
instances, or data in a database table and in
a file on disk, though this is not yet implemented.
(The next post will discuss
tdda difffurther.) - In the case of the API, this can also include in-memory data structures such as data frames.
- In the case of the command-line tool this is two datasets on
disk (flat files, parquet files etc.). It might also be possible
to compare two database tables, in the same or different RDBMS
instances, or data in a database table and in
a file on disk, though this is not yet implemented.
(The next post will discuss
-
Provide Metadata Format Conversions. Provide mechanisms for converting between different library-specific flat-file parameters and tdda's
tdda.serialformat, as well as between thetdda.serialformat,csvw, and (perhaps)frictionless. - Generate Validation Statistics and Validate using them. (Potentially) write additional data for a concrete dataset that can be used for further validation that it has been read correctly, e.g. summary statistics, checksums etc.
Discussion
The usual observation when proposing something new like this is that the last thing the world needs is another “standard”. As Randall Munro puts it: (https://imgs.xkcd.com/927):

In this case, however, I don't think there are all that many
recognized ways of describing flat-file formats. I was involved in one
(the .fdd flat-file description data format) while at Quadstone, and
I currently use the XMD format above at Stochastic Solutions, but
pretty-much no one else does. While working with a friend, Neil
Skilling, he ran across the CSVW standard,
developed under the auspices of W3C, and that led to my finding the
Python frictionless project.
At first I thought one of those might be the solution I was looking
for, but in fact they have goals and desgins that are different enough
that they don't quite fulfill the most important goals for
tdda.serial, as impressive as both projects are.
Reluctantly, therefore, I began working on tdda.serial,
which aims to interoperate with and support CSVW, (and to some extent,
frictionless), but also to handle other cases.
The biggest single difference between the focus of tdda.serial
and the CSVW is that tdda.serial is primarily concerned with documenting
a format that might be used by many flat files (different concrete
datasets sharing the same sttructure and formatting) whereas CSVW
is primarily concerned with documenting either
a single specific CSV file or a specific collection of CSV files,
usually each having different structure. This seems like a rather
subtle difference, but in fact turns out to be quite consequential.
Here's the first example CSVW file from csvw.org:
{
"@context": ["http://www.w3.org/ns/csvw", {"@language": "en"}],
"tables": [{
"url": "http://opendata.leeds.gov.uk/downloads/gritting/grit_bins.csv",
"tableSchema": {
"columns": [
{
"name": "location",
"datatype": "integer"
},
{
"name": "easting",
"datatype": "decimal",
"propertyUrl": "http://data.ordnancesurvey.co.uk/ontology/spatialrelations/easting"
},
{
"name": "northing",
"datatype": "decimal",
"propertyUrl": "http://data.ordnancesurvey.co.uk/ontology/spatialrelations/northing"
}
],
"aboutUrl": "#{location}"
}
}],
"dialect": {
"header": false
}
}
Notice that the CSVW file caters for multiple CSV files (a list of tables
in the tables element), and that the location of the table is provided
as a URL (which is a required element in CSVW).
In the context of CSV on the web, this makes complete sense.
It's specified as being URL, but can be a file: URL, or
a simple path. One convention, fora CSVW file documenting a single
dataset, seems to be that the metadata for
grit_bins.csv is stored in grit_bins-metadata.json in the same
directory as the CSV file itself (locally, or on the web).
What is significant, however, is that this establishes either a one-to-one relationship between CSV files and CSVW metadata files or, if the CSVW file contains metadata about several files, a one-to-one relationship between CSVW files and metadata tables in a CSVW file. Here, for example, is Example 5 from the CSVW Primer:
{
"@context": "http://www.w3.org/ns/csvw",
"tables": [{
"url": "countries.csv"
}, {
"url": "country-groups.csv"
}, {
"url": "unemployment.csv"
}]
}
The metadata “knows” the data file
(or data files) that it describes. In contrast, the main concern of
tdda.serial is to describe a format and structure that might well be
used for many specific (“concrete”) flat files. The relationship is
almost reversed as shown here:
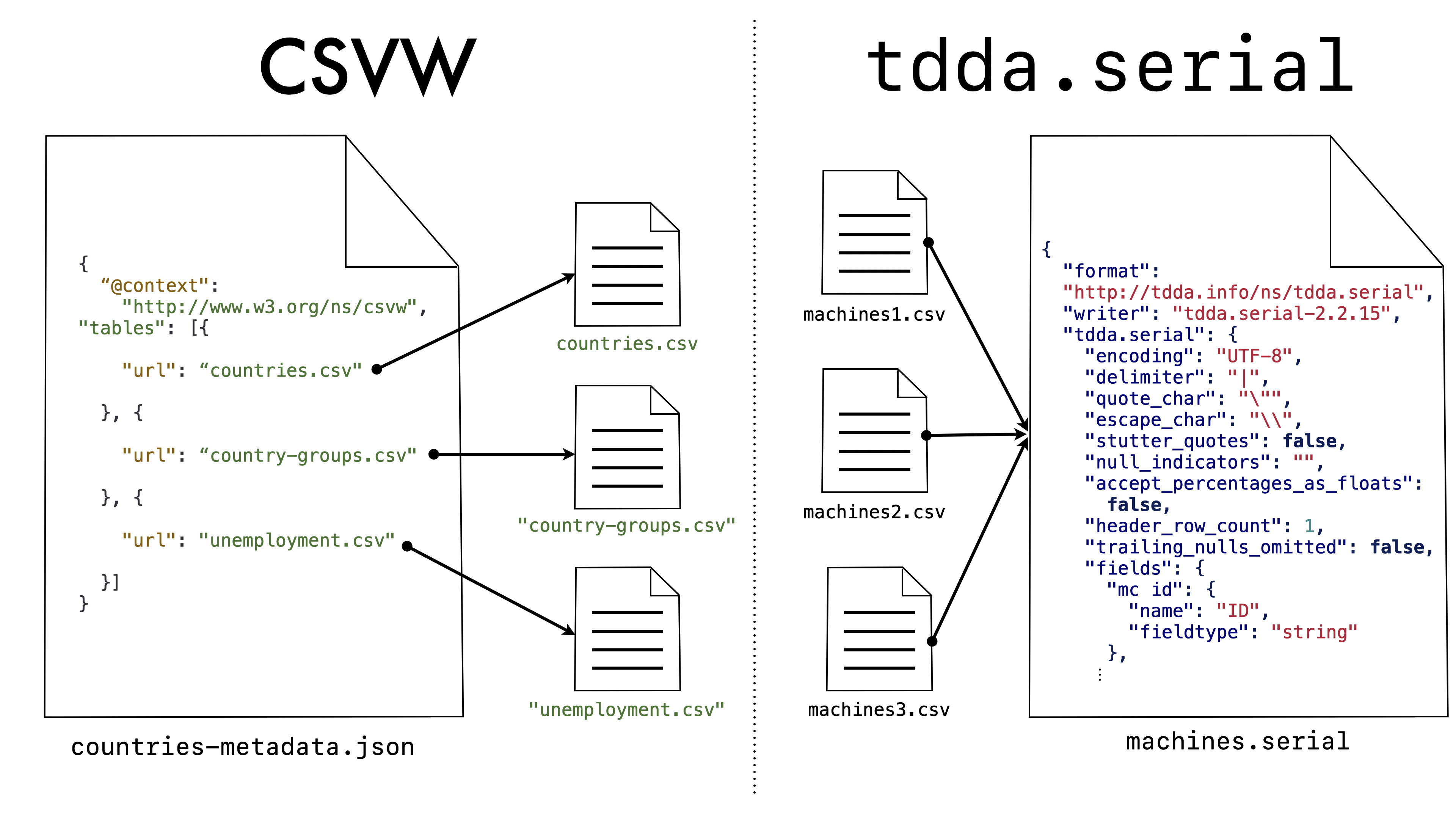
Even though the URL (url) is a mandatory parameter in CSVW, there is
nothing to prevent us from taking a CSVW file (particularly one
describing a single table) and using its metadata to define a format
to be used with other flat files. In doing, however, we would clearly
be going against the grain of the design of CSVW. As an example
of how it then does not quite fit, sometimes we want the metadata to
describe exactly the fields in the data, and other times we want it to
be a partial specification. In the XMD file, there are explicit
parameters to say whether or not extra fields are allowed, and whether
all fields are required. In the case of the tdda.serial file, we use
a list of fields when we are describing all the fields allowed and
required in a flat file, and a dictionary when we are providing
information only on a subset, not necessarily in order.4
This sort of flexibility is harder in CSVW, which always uses a list
to specify the fields. I could propose and use extensions, or try to
get extensions added to the standard, but the former seem undesirable,
and the latter hard an unlikely. (It does not look as if there have
been and revisions to CSVW since 2022.)
There are, in fact, many details of CSVW that are problematical
for even the first two libaries I've looked at (Pandas and Polars),
so unfortunately I think something different is needed.
Library-specific Support in tdda.serial
Another goal for tdda.serial is that it should be
useful even for people who are only using a single library—e.g.
Pandas. In such cases, there is typically a function or
method for writing CSV files (pandas.DataFrame.to_csv), and another for
reading them (pandas.read_csv). Both typically have many
optional arguments, and in keeping with Postel's Law (the
Robustness Principle),
they typically have more flexibility in read formats than in write formats.
In the case of Pandas, the read function's signature is:
pandas.read_csv(
filepath_or_buffer, *, sep=<no_default>,
delimiter=None, header='infer', names=<no_default>, index_col=None,
usecols=None, dtype=None, engine=None, converters=None,
true_values=None, false_values=None, skipinitialspace=False,
skiprows=None, skipfooter=0, nrows=None, na_values=None,
keep_default_na=True, na_filter=True, verbose=<no_default>,
skip_blank_lines=True, parse_dates=None,
infer_datetime_format=<no_default>, keep_date_col=<no_default>,
date_parser=<no_default>, date_format=None, dayfirst=False,
cache_dates=True, iterator=False, chunksize=None,
compression='infer', thousands=None, decimal='.',
lineterminator=None, quotechar='"', quoting=0, doublequote=True,
escapechar=None, comment=None, encoding=None,
encoding_errors='strict', dialect=None, on_bad_lines='error',
delim_whitespace=<no_default>, low_memory=True, memory_map=False,
float_precision=None, storage_options=None,
dtype_backend=<no_default>
)
(49 parameters), while the write method's signature is:
DataFrame.to_csv(
path_or_buf=None, *, sep=',', na_rep='',
float_format=None, columns=None, header=True, index=True,
index_label=None, mode='w', encoding=None, compression='infer',
quoting=None, quotechar='"', lineterminator=None, chunksize=None,
date_format=None, doublequote=True, escapechar=None, decimal='.',
errors='strict', storage_options=None
)
(22 parameters).
The tdda library's command-line tools allow a tdda.serial
specification to be converted to parameters for pandas.read_csv,
returning them as a dictionary that can be passed in using
**kargs. It can also generate python
code to do the read using pandas.read_csv
or directly perform the read, saving the result to parquet.
Similarly, the library can take a set of arguments for DataFrame.to_csv
and create a tdda.serial file describing the format used (or
write the data and metadata together).
For a user working with a single library, however, converting to and from
tdda.serial's metadata description might be unnecessarily cumbersome and
may work imperfectly. This is because different libraries represent
data differently, and are based on slighlty different conceptions of CSV files.
While I am going to make some effort to allow tdda.serial universal,
it is likely that there will always be some cases in which there is
a loss of fidelity moving between any specific library's arguments
and the .serial representation.
For these reasons, the tdda library also supports directly writing
arguments for a given library. That is why the tdda.serial metadata
description is one level down inside the tdda.serial file, under
a tdda.serial key. It is also possible to have sections for
pandas.read_csv, polars.read_csv with exactly the arguments
they need.
-
The functionality used on this post is not in the release version of the tdda library, but is there on a branch called
detectreport, so can be accessed if anyone it particulary keen. ↩ -
In fact, in writing this post, I updated the previous one to use a slightly more sensible example that previously; this is the new, slightly more useful example. ↩
-
CSV is not a very suitable format for perfect round-tripping of data for reasons including numeric rounding, multiple types for the same data, and equivalent representations such as string and categoricals. Even using a typed format such as parquet, some of these details may change on round-tripping and most software needs a library-specific format in order to achieve perfect fidelity when serializing and deserializing data. ↩
-
This precise mechanism may change, but it is important for
tdda.serial's purpose that is supports both full and partial field schema specification. ↩